How to create DVH of small RoI?
DVH in small RoI appears as staircases. We will explain how to improve the histogram calculation.
The following instructions will load the patient data into memory:
library (espadon)
# patient loading
pat.dir <- choose.dir ()
pat <- load.patient.from.dicom(pat.dir)
S <- load.obj.data (pat$rtstruct[[1]])
D <- load.obj.data (pat$rtdose[[1]]) We suppose that:
- D is an espadon object of modality rtdose, with a D$patient.orientation equal to c (1, 0, 0, 0, 1, 0). This means that the orientation of the cutting planes is the same as that of the patient. The volume of D voxel is 3x3x3 mm3= 0.027cm3 (= abs(prod(D$dxyz))).
- S is an espadon object of of modality rtstruct containing the RoI “amygdala”, and with the same frame of reference as D. The volume of the chiasma is 1.38 cm3 (= S$roi.info$vol[S$roi.info$roi.pseudo == ” amygdala”])
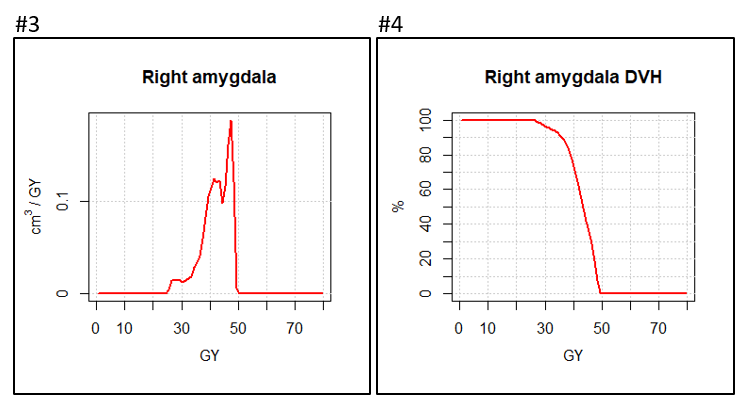
The volume of the amygdala contains about 51 voxels.
If the histograms in the chiasm is calculated without oversampling, this generates staircases on DVH.
# Histogram calculation
roi.idx <- select.names(S$roi.info$roi.pseudo, roi.sname = "amygda")[1]
h <- histo.from.roi (D , S, roi.idx = roi.idx, breaks = seq (0, 80, 1),
alias = S$roi.info$name[roi.idx])
display.dV_dx (h, MC.plot = T, col = "#ff0000", lwd = 2) #1
DVH <- histo.DVH (h, alias = paste(S$roi.info$name[roi.idx], "DVH"))
display.DVH.pc (DVH, col = "#ff0000", lwd=2) #2 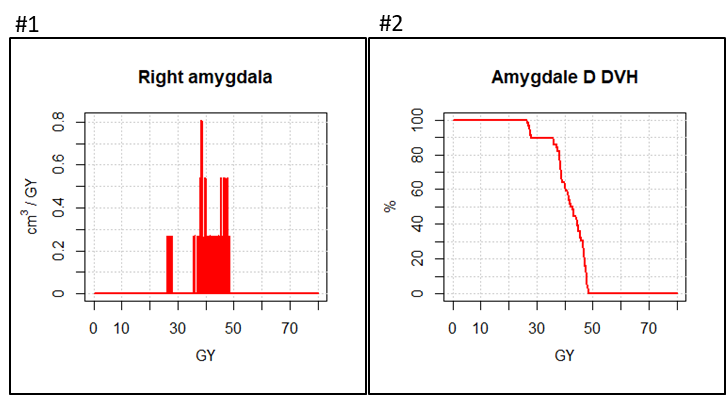
The following instructions explain how to resample volume D, to improve the calculation
# First limit the compute volume
nesting.D <-nesting.roi (D, struct = S, roi.idx = roi.idx, xyz.margin = c (10, 10, 10))
# Create an oversampled volume resample.D
n <- 10
resample.D <- vol.oversampling(nesting.D, fact.ijk = c(n,n,1))
# Create and display the histogram
h <- histo.from.roi(resample.D , S, roi.idx = roi.idx, breaks = seq (0, 80, 1),
alias = S$roi.info$name[roi.idx])
display.dV_dx(h, col = "#ff0000", lwd = 2) #3
DVH <- histo.DVH(h, alias = paste(S$roi.info$name[roi.idx], "DVH"))
display.DVH.pc(DVH, col = "#ff0000", lwd = 2) #4