Get started
2D Display
Visualization of imaging volumes is essential for controlling the loaded DICOM objects. The espadon package allows this display, with the possibility of using the color palettes of your choice.
The display.kplane() function displays a slice plane of the imaging volume in the slice plane geometric frame of reference.
CT <- load.obj.from.dicom(ct_dicom_file_path, data = TRUE)
display.kplane (CT, k =10, col = pal.RVV(1000),
breaks = seq(-1000, 1000, length.out = 1001),
ord.flip = TRUE, interpolate = TRUE) 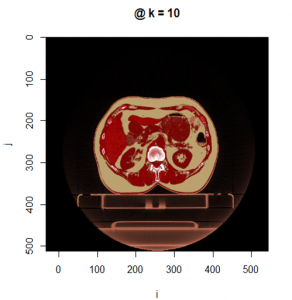
[1] Silverstein JC, Parsad NM, Tsirline V (2008). “Automatic perceptual color map generation for realistic volume visualization.” Journal of Biomedical Informatics, 41(6), 927-935. ISSN 1532-0464, doi: 10.1016/j.jbi.2008.02.008.
The display.plane() function displays transverse, frontal or sagittal views in the patient’s frame of reference. It allows the superposition of images and contours.
# pat.dir is the patient directory containing the DICOM files for a MR, an RT-dose and an RT-struct, and a REG file to link RT-dose and MR frames of reference.
patient <- load.patient.from.dicom(pat.dir, data = TRUE)
D <- load.obj.data(pat.f$rtdose[[1]])
S <- load.obj.data(pat.f$rtstruct[[1]])
MR <- load.obj.data(pat.f$mr[[1]])
# Maximum dose coordinates
Max.dose.coord <- get.xyz.from.index (which(abs(D$vol3D.data-D$max.pixel)<1e-3), D)[1,]
# Display in the frame of reference of RT-dose
display.plane(bottom = MR, top = D, struct = S, view.type = "trans",
view.coord = Max.dose.coord[3], display.ref = D$ref.pseudo,
T.MAT = pat.f$T.MAT)
display.plane(bottom = MR, top = D, struct = S, view.type = "front",
view.coord = Max.dose.coord[2], display.ref = D$ref.pseudo,
T.MAT = pat.f$T.MAT)
display.plane(bottom = MR, top = D, struct = S, view.type = "sagi",
view.coord = Max.dose.coord[1], display.ref = D$ref.pseudo,
T.MAT = pat.f$T.MAT)
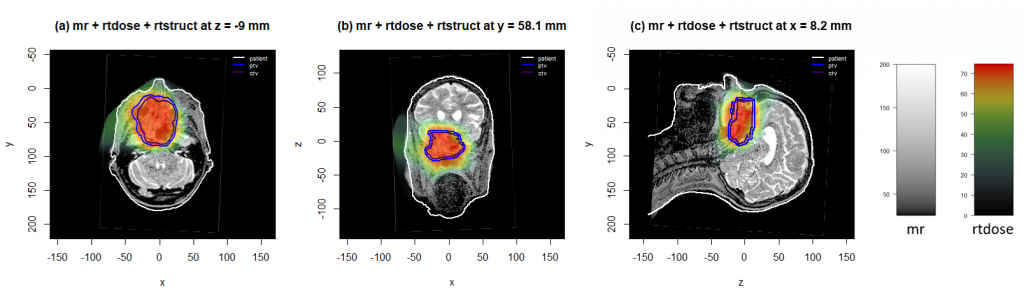
It is also possible to use the plot() function, which can display objects of class volume (for CT, MR, RT-dose for example), objects of class struct (for RT-struct) and those of class mesh.
plot (D, view.type = "trans", view.coord = Max.dose.coord[3], col =pal.rainbow(255))
plot (S, view.type = "trans", view.coord = Max.dose.coord[3], add = TRUE)
