Get started
Binary objects
Binary objects are volumes whose voxel values are either TRUE, FALSE or NA (i.e. undetermined). They act as a mask, and are used to select only the desired part of a volume.
The example below show how to select the brain voxels described in the rtstruct object.
nesting.MR <- nesting.roi (MR, S, roi.sname = "brain", T.MAT = pat$T.MAT, vol.restrict = TRUE,
xyz.margin = c(20, 20, 20)) # to reduce the computation time
bin.brain <- bin.from.roi (nesting.MR, struct = S, roi.sname = "brain", T.MAT = pat$T.MAT)
MR.brain <- vol.from.bin (nesting.MR, bin.brain) 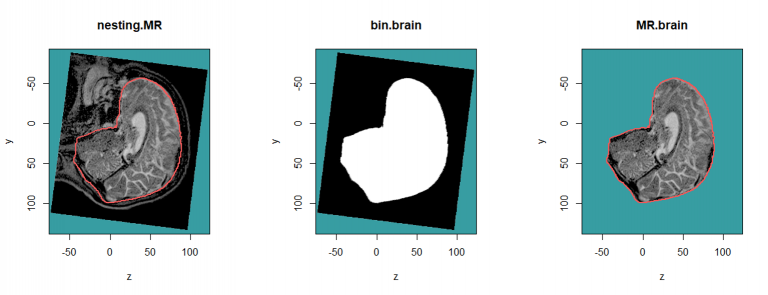
The bin.from.vol function creates binary volumes by selecting voxels that are inside or outside an interval [min, max]. The example below selects voxels with a value greater than 150.
bin.min.150 <- bin.from.vol (MR.brain, min = 150)
display.plane (bin.min.150, view.coord = 0, view.type = "sagi", main = "bin.min.150",
bg = "#379DA2", legend.plot = FALSE, sat.transp = TRUE, interpolate = TRUE) 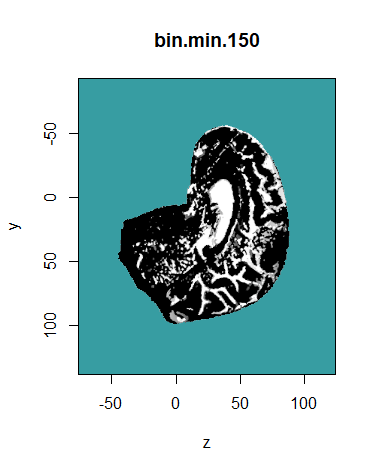
The usual functions on binary volumes, such as inversion (bin.inversion), intersection (bin.intersection), union (bin.sum) and subtraction (bin.subtraction), have been implemented in the espadon package.

Other image processing functions are available:
- bin.erosion decreases the volume of a radius r
- bin.dilation increases the volume of a radius r
- bin.closing is usefull for filling holes that are smaller than the radius and merging two shapes close to each other.
- bin.opening is usefull for removing volumes that are smaller than the radius and smoothing shapes.
- bin.clustering groups and labels TRUE voxels that have a 6-connectivity (i.e. sharing a common side).
These functions can be used to separate multiple volumes in the binary selection, as shown below:
bin.smooth <- bin.opening (bin.min.150, radius = 1.5))
bin.cluster <- bin.clustering (bin.smooth)
# Creation of ventricle contours
bin.ventricle <- bin.from.vol (bin.cluster, min = 1, max = 1)
S.ventricle <- struct.from.bin (bin.ventricle , roi.name = "ventricle", roi.color = "#FF0000" )
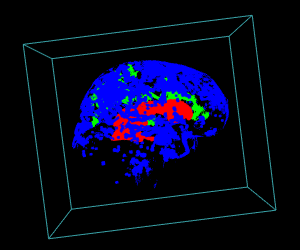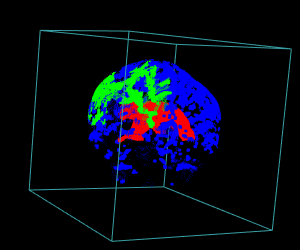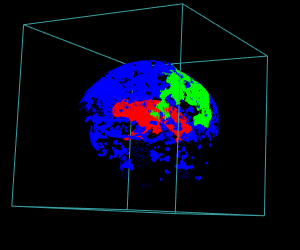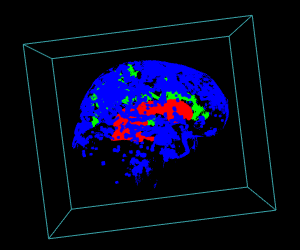
# Display of the first 3 largest sub-volumes, and ventricle contours
library (rgl)
bg3d ("black")
par3d (userMatrix = matrix (c(0, 0, -1, 0, -1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1), ncol = 4),
windowRect = c(0, 50, 300, 300), zoom = 0.7)
display.3D.stack(bin.cluster, k.idx = bin.cluster$k.idx, col = c ("#00000000", rainbow (3, alpha = 1)),
breaks = seq (0, 3, length.out = 5), border = FALSE, ktext = FALSE)
play3d (spin3d (rpm = 4), duration = 15)
bg3d ("black")
par3d (userMatrix = matrix (c(0, 0, -1, 0, -1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1), ncol = 4),
windowRect = c(0, 50, 300, 300), zoom = 1)
display.3D.contour (S.ventricle)
play3d (spin3d (rpm = 4), duration = 15)